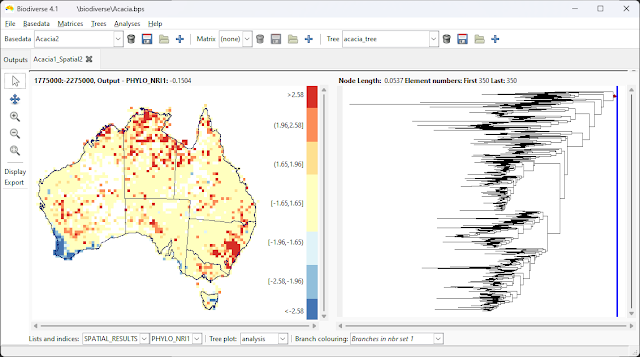
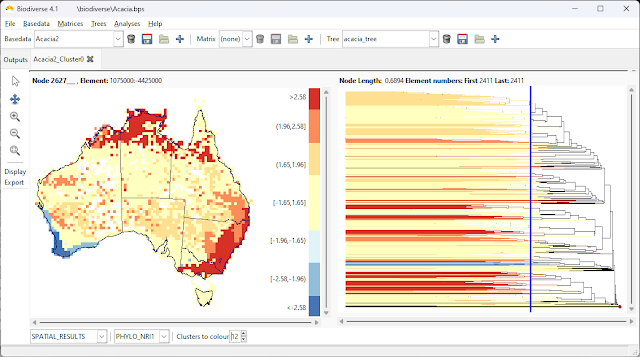
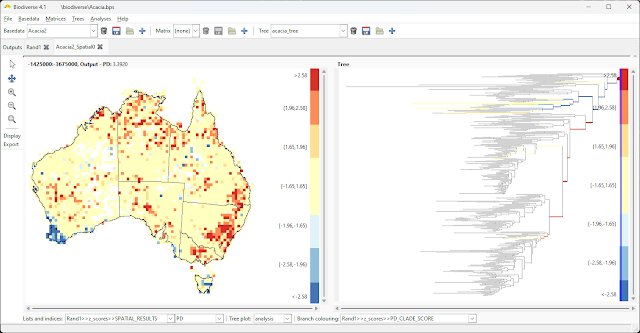
2023 is now in full swing, so here is a list of publications from 2022 that used Biodiverse.
-
Amaral, D.T., Bonatelli, I.A.S., Romeiro-Brito, M., Moraes, E.M. and
Franco, F.F. (2022) Spatial patterns of evolutionary diversity in
Cactaceae show low ecological representation within protected areas. Biological Conservation, 273, 109677.
-
Ávila-González, H., González-Gallegos, J.G., Munguía-Lino, G. &
Castro-Castro, A. (2022) The genus Sisyrinchium (Iridaceae) in Sierra
Madre Occidental, Mexico: A new species, richness and distribution. Systematic Botany, 47, 319-334.
-
Carter, B. E., Misiewicz, T. M. & Mishler, B. D. (2022). Spatial
phylogenetic patterns in the North American moss flora are shaped by
history and climate. Journal of Biogeography, 49, 1327-1338.
-
Chen, K., Khine, P.K., Yang, Z. and Schneider, H. (2022) Historical
plant records enlighten the conservation efforts of ferns and
Lycophytes’ diversity in tropical China, Journal for Nature Conservation, 68, 126197.
-
Contreras-Medina, R., García-Martínez, A. I., Ramírez-Martínez, J.
C., Espinosa, D., Balam-Narváez, R., and Luna-Vega, I. (2021).
Biogeographic analysis of ferns and lycophytes in Oaxaca: A Mexican
beta-diverse area. Botanical Sciences, 100, 204-222.
-
Fernandes, N.B.G., de Menezes Yazbeck, G. & Milward-de-Azevedo,
M.A. (2022) Taxonomic diversity of Passifloraceae sensu stricto along
altitudinal gradient and on Serra dos Órgãos mountain slopes in
southeastern Brazil. Rodriguésia, 73, e00702021.
-
Gosper C.R., Percy-Bower J.M., Byrne M., Llorens T.M. & Yates
C.J. (2022) Distribution, Biogeography and Characteristics of the
Threatened and Data-Deficient Flora in the Southwest Australian
Floristic Region. Diversity, 14, 493.
-
Griffiths, D. (2022). Do the drivers and levels of isolation in fish
faunas differ across Atlantic and Pacific drainages in the Americas? Journal of Biogeography, 49, 930-941.
-
Gutiérrez-Rodríguez, B.E., Guevara, R., Angulo, D.F. et al. (2022)
Ecological niches, endemism and conservation of the species in Selenicereus (Hylocereeae, Cactaceae). Brazilian Journal of Botany, 45, pages 1149–1160.
-
Gutiérrez–Rodríguez, B.E., Vásquez–Cruz, M. and Sosa, V. (2022)
Phylogenetic endemism of the orchids of Megamexico reveals complementary
areas for conservation. Plant Diversity, 44, 351-359.
-
Kong, H., Condamine, F.L., Yang, L., Harris, A.J., Feng, C., Wen, F.
and Kang, M. (in press) Phylogenomic and macroevolutionary evidence for
an explosive radiation of a plant genus in the Miocene. Systematic Biology, 71, 589–609.
-
Moreira-Muñoz, A, Palchetti, V.A., Morales-Fierro, V., Duval, V.S.,
Allesch-Villalobos, R., & González-Orozco, C.E. (2022) Diversity and
Conservation Gap Analysis of the Solanaceae of Southern South America.
Frontiers in Plant Science, 13.
-
Murillo-Pérez, G., Rodríguez, A., Sánchez-Carbajal, D., Ruiz-Sanchez,
E., Carrillo-Reyes, P., Munguía-Lino, G. (2022) Spatial distribution of
species richness and endemism of Solanum (Solanaceae) in Mexico. Phytotaxa 558, 147–177
-
Olivares-Juárez, M.I., Burgos-Hernández, M. and Santiago-Alvarádo, M.
(2022) Patterns of Species Richness and Distribution of the Genus Laelia s.l. vs. Laelia s.s. (Laeliinae: Epidendroideae: Orchidaceae) in Mexico: Taxonomic Contribution and Conservation Implications. Plants, 11:2742.
-
Paz, A., Silva, A.S. & Carnaval, A. (2022) A framework for
near-real time monitoring of diversity patterns based on indirect remote
sensing, with an application in the Brazilian Atlantic rainforest. PeerJ, 10:e13534.
-
Rivera-Martínez, R., Ramírez-Morillo, I.M., De-Nova, José A.,
Carnevali, G., Pinzón, J.P., Romero-Soler, K.J. & Raigoza, N. (2022)
Spatial phylogenetics in Hechtioideae (Bromeliaceae) reveals recent
diversification and dispersal. Botanical Sciences, 100, 692-709.
-
Silva, D.C., Oliveira, H.F.M., Zangrandi, P.L. and Domingos, F.M.C.B.
(2022) Flying Over Amazonian Waters: The Role of Rivers on the
Distribution and Endemism Patterns of Neotropical Bats. Frontiers in Ecology and Evolution, 10:774083.
-
Wang, Q., Huang, J., Zang, R., Li, Z. and El-Kassaby, Y. A. (2022).
Centres of neo- and paleo-endemism for Chinese woody flora and their
environmental features. Biological Conservation, 276, 109817.
-
Yang, X., Qin, F., Xue, T., Xia, C., Gadagkar, S. R., & Yu, S.
(2022). Insights into plant biodiversity conservation in large river
valleys in China: A spatial analysis of species and phylogenetic
diversity. Ecology and Evolution, 12, e8940.
-
Yang, X., Zhang, W., Qin, F., et al. (2022). Biodiversity priority areas and conservation strategies for seed plants in China. Frontiers in Plant Science, 13, 962609.
-
Zhang, W., Bussmann, R.W., Li, J., Liu, B., Xue, T., Yang, X., Qin,
F., Liu, H. and Yu, S. (2022) Biodiversity hotspots and conservation
efficiency of a large drainage basin: Distribution patterns of species
richness and conservation gaps analysis in the Yangtze River Basin,
China. Conservation Science and Practice, 4, e12653.
-
Zhang, Y., Qian, L., Chen, X., Sun, L., Sun, H. and Chen, J. (2022)
Diversity patterns of cushion plants on the Qinghai-Tibet Plateau: a
basic study for future conservation efforts on alpine ecosystems. Plant Diversity, 44, 231-242.
-
Zhang, X.X, Ye, J.F., Laffan, S.W., Mishler, B.D., Thornhill, A.H.,
Lu, L.M. et al. (2022) Spatial phylogenetics of the Chinese angiosperm
flora provides insights into endemism and conservation. Journal of Integrative Plant Biology, 64, 105-117.
-
Zhao, R., Xu, S., Song, P., Zhou, X, Zhang, Y. and Yuan, Y. (2022)
Distribution patterns of medicinal plant diversity and their
conservation in the Qinghai-Tibet Plateau. Biodiversity Science, 30, 21385.