Biodiverse version 2.0 is now available. It can be accessed from https://github.com/shawnlaffan/biodiverse/wiki/Downloads
The summary of changes since version 1.1 is at https://github.com/shawnlaffan/biodiverse/wiki/ReleaseNotes#version-20
Blog posts describing much of the new functionality can be accessed at http://biodiverse-analysis-software.blogspot.com.au/
The available binary versions work only on 64 bit computers, but windows, linux and mac operating systems are all supported.
Installation instructions are at https://github.com/shawnlaffan/biodiverse/wiki/Installation
Please report any errors or suggested improvements. You can use the
email list at https://groups.google.com/forum/#!forum/biodiverse-users or the project issue tracker at
https://github.com/shawnlaffan/biodiverse/issues
Shawn Laffan
15-Nov-2017
Wednesday, 15 November 2017
Tuesday, 19 September 2017
Log scale your map displays
In Biodiverse version 2 you will be able to log scale the colour stretch on the map. This is a pretty minor change in some ways, but does allow much better visualisations of results when the distributions are highly left skewed.
The Spatial and Cluster tabs have a legend option to allow you to turn it on or off.
The View Labels tab currently switches between log and linear scaling depending on the maximum value and the maximum possible value. It will use log scaling when there are more than 20 labels to highlight or the the maximum value across highlighted cells is less than 0.8 times the number of selected labels. Otherwise it will use a linear scaling (so if there are 20 selected labels, but no cell contains more than 16 then it will log scale the display).
Here is a video to demonstrate the process. The first part shows a matrix visualisation of a Region Grower analysis (a simple pair-wise complementarity analysis, in this case using richness as the index). It is much easier to see the spatial patterns when the colour stretch is log scaled. The second shows a spatial analysis. In this case the log scaling does not make too much difference. The third shows the view labels tab. Note that the log scaling makes it much easier to see an entire clade on a tree, as previously the colours would have been washed out.
The Spatial and Cluster tabs have a legend option to allow you to turn it on or off.
The View Labels tab currently switches between log and linear scaling depending on the maximum value and the maximum possible value. It will use log scaling when there are more than 20 labels to highlight or the the maximum value across highlighted cells is less than 0.8 times the number of selected labels. Otherwise it will use a linear scaling (so if there are 20 selected labels, but no cell contains more than 16 then it will log scale the display).
Here is a video to demonstrate the process. The first part shows a matrix visualisation of a Region Grower analysis (a simple pair-wise complementarity analysis, in this case using richness as the index). It is much easier to see the spatial patterns when the colour stretch is log scaled. The second shows a spatial analysis. In this case the log scaling does not make too much difference. The third shows the view labels tab. Note that the log scaling makes it much easier to see an entire clade on a tree, as previously the colours would have been washed out.
Shawn Laffan
19-Aug-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
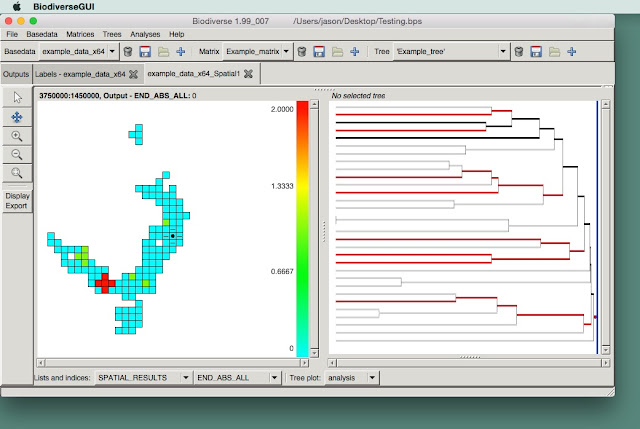
Visualise spatial analysis results on the tree
Many of the indices in Biodiverse allow users to see what the relative contribution of branches on the tree are to one or more indices. For example, one can see what the weighted branch lengths are in a phylogenetic endemism analysis, or see what the relative loss of phylogenetic diversity would be if a branch was lost from a sample.
Previously the only way to inspect these was to use a popup window by control-clicking on a cell. This is useful, but rapidly becomes difficult when there are many cells to explore.
In version 2 of Biodiverse, users can now visualise these values on the tree as the mouse is hovered over cells.
This builds on the turnover displays that were added a while ago.
Rather than a long blog post, it is probably best to simply show a video. In this example, a set of indices are calculated for the Acacia data set described in several publications (see the full list here and search for Acacia).
Note how the display is log scaled by default, but linear scaling can be used if preferred. The default colour scheme uses the rainbow-ish default, as in the map display, but other schemes can also be used. Users can also hide the legend if it gets in the way.
Shawn Laffan
19-Aug-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Export lists to Newick format
In version 2 of Biodiverse you can now export any lists that are stored on a tree to the Nexus format.
A useful example of this is when you run calculations for each node of a cluster tree, thereby using the terminal nodes (branches) below each branch as the neighbourhood units in a spatial analysis. An example of this in practice is in González-Orozco et al. (2014) where the environmental parameters of each biogeographic region were summarised (see table 4 in that paper).
In a previous post I described how you can export the colours from the Biodiverse display to Nexus format to more easily generate figures for publication. Now you can also export the values of the results to Nexus, which means you can use them for further analysis or alternate display methods using tools such as FigTree or Mesquite.
Below are images showing the general process in practice.
Note that the Nexus format does not support a hierarchy of names, so the lists need to be flattened. Given that it is possible to have items with the same value within different lists in Biodiverse (e.g. for list indices that generate a result per label in a set), the exported names use the list name followed by the item name, joined by two underscores e.g. SPATIAL_RESULTS__ENDC_CWE.
A useful example of this is when you run calculations for each node of a cluster tree, thereby using the terminal nodes (branches) below each branch as the neighbourhood units in a spatial analysis. An example of this in practice is in González-Orozco et al. (2014) where the environmental parameters of each biogeographic region were summarised (see table 4 in that paper).
In a previous post I described how you can export the colours from the Biodiverse display to Nexus format to more easily generate figures for publication. Now you can also export the values of the results to Nexus, which means you can use them for further analysis or alternate display methods using tools such as FigTree or Mesquite.
Below are images showing the general process in practice.
Note that the Nexus format does not support a hierarchy of names, so the lists need to be flattened. Given that it is possible to have items with the same value within different lists in Biodiverse (e.g. for list indices that generate a result per label in a set), the exported names use the list name followed by the item name, joined by two underscores e.g. SPATIAL_RESULTS__ENDC_CWE.
 |
| Setting up a cluster analysis where a set of endemism indices will be calculated for each branch in the cluster tree. |
 |
| Displaying the results, in this case the Corrected Weighted Endemism (CWE) for the set of branches intercepted by the blue slider bar on the dendrogram |
 |
| The exported tree, but with default display settings. |
 |
| And now with thicker branches, and the colours set to show the CWE results using a divergent red-blue colour scheme. |
 |
| And with the Richness index plotted using the HSB spectrum. |
And here is a video of the process in Biodiverse (partly as a test to see if it works):
Shawn Laffan
19-Aug-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Monday, 21 August 2017
Copy selected records to the clipboard
In version 1, users were able to copy the selected labels from the View Labels tab so they could paste it into other applications (or elsewhere in Biodiverse).
In version 2, users will also be able to copy all columns for the selected records.
This means you can easily get a list of species and the associated summary statistics (variety, sample count and redundancy), plus any label properties that have been attached to the data.
In version 2, users will also be able to copy all columns for the selected records.
This means you can easily get a list of species and the associated summary statistics (variety, sample count and redundancy), plus any label properties that have been attached to the data.
 |
| Users can now copy selected records to the clipboard, not just the labels. |
 |
| Records can be pasted into any application, for example spreadsheets. |
 |
| And if you are an R user then you can use the read.table() function. (set row.names=1 to use the labels as the row names) |
Shawn Laffan
21-Aug-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Thursday, 27 April 2017
Biodiverse now works on Macs
Thanks to some sterling work by Jason Mumbulla we now have a native version of Biodiverse for Macintosh computers.
Installation is by dmg file, so follows the same approach as most mac installations.
The interface is somewhat "old school" (think 1998), but all the functionality is in place and works (see screenshots below).
Download links are at https://purl.org/biodiverse/wiki/Downloads (at the time of writing there is only a development release), with installation instructions at https://purl.org/biodiverse/wiki/Installation.
Shawn Laffan
27-Apr-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Wednesday, 26 April 2017
Biodiverse now exports tree branch colours
Biodiverse version 2 will allow you to export the tree branch colours. This means you can now get your tree colours to the point that you are happy with them, and export them to another program such as FigTree to generate figures for a publication.
Colours can be exported with the Nexus and tabular Tree formats. If you use the tabular format then there will be a column in the output table called Colour. For the Nexus format, the colours are stored in the comments block for each branch.
A few examples are below.
The colouring works for anything that is applied to the tree, so you can also export continuous palettes when you have calculations per node (branch).
Kudos to Luke Fitzpatrick for getting it working.
Shawn Laffan
Colours can be exported with the Nexus and tabular Tree formats. If you use the tabular format then there will be a column in the output table called Colour. For the Nexus format, the colours are stored in the comments block for each branch.
 |
| Click on the "Export colours" checkbox to export the colours in a Nexus file. |
A few examples are below.
 |
| View labels tab with a set of labels highlighted on the tree. |
 |
| The selection colours displayed in FigTree. |
 |
| A cluster analysis with six clusters coloured. |
 |
| The tree in FigTree with the six clusters coloured. |
 |
| A user defined selection. |
 |
| The same user defined selection in FigTree. |
The colouring works for anything that is applied to the tree, so you can also export continuous palettes when you have calculations per node (branch).
 |
| PD calculated for each cluster node (branch). |
 |
| And the same PD colouring now in FigTree. |
Kudos to Luke Fitzpatrick for getting it working.
Shawn Laffan
26-Apr-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Matching spatial, tree, matrix and property data is now easier
One of the biggest bugbears when using Biodiverse has been matching the names between the spatial data and any tree, matrices, group properties and label properties.
Biodiverse uses an exact matching scheme to link tree branch names with Basedata labels (e.g. species). If there is even a single character different then the two sets will not match, and analyses either do not run or are incomplete.
The standard approach used in Biodiverse to ensure the data match is for users to provide a remap table that "maps" labels from one data set to another (e.g. make the tree names match the basedata). However, experience suggest that users are often unsure what columns should be used for what (and sometimes how many), and that generation of the remap file is more difficult than desired.
Biodiverse version 2 has a greatly simplified remap process that can generate possible matches automatically. Any object can have its element names remapped at import, as before, but now there are also options under the Basedata, Tree and Matrix menus. Better yet, there is also a centralised interface that can be accessed under the file menu.
In the centralised remap interface, one can match any object to any other object in the project, as well as to names loaded from a file. If an object is chosen then Biodiverse will search the sets of names in each object to find possible matches. The User defined from file is the conventional process, where the file needs to define the set of input and remapped columns. The Auto from file allows users to load a list of names which will be searched to find possible matches, in the same way as it searches other objects.
The search process uses a fuzzy text matching scheme, as well as searching for differences purely in punctuation or in quoting characters. The minimum acceptable distance option allows control over how different possible matches can be. If a very large value is used then anything can match anything else, which is unlikely to be useful.
The interface for the automatic remaps is as below. There are panes listing exact matches, non-matches (i.e. differences were too great), punctuation matches and possible typos. Users can choose to ignore any of these sets, and also select subsets within the sets if, for example, there are false matches.
Users can also choose to export the remap to a file to re-use later on (or inspect for possible problems) using the Export remap to file button. A copy of the remap can also be sent to the clipboard for direct use in spreadsheets, text editors and the like.
Kudos to Luke Fitzpatrick who got it all working.
Biodiverse uses an exact matching scheme to link tree branch names with Basedata labels (e.g. species). If there is even a single character different then the two sets will not match, and analyses either do not run or are incomplete.
The standard approach used in Biodiverse to ensure the data match is for users to provide a remap table that "maps" labels from one data set to another (e.g. make the tree names match the basedata). However, experience suggest that users are often unsure what columns should be used for what (and sometimes how many), and that generation of the remap file is more difficult than desired.
Biodiverse version 2 has a greatly simplified remap process that can generate possible matches automatically. Any object can have its element names remapped at import, as before, but now there are also options under the Basedata, Tree and Matrix menus. Better yet, there is also a centralised interface that can be accessed under the file menu.
In the centralised remap interface, one can match any object to any other object in the project, as well as to names loaded from a file. If an object is chosen then Biodiverse will search the sets of names in each object to find possible matches. The User defined from file is the conventional process, where the file needs to define the set of input and remapped columns. The Auto from file allows users to load a list of names which will be searched to find possible matches, in the same way as it searches other objects.
The search process uses a fuzzy text matching scheme, as well as searching for differences purely in punctuation or in quoting characters. The minimum acceptable distance option allows control over how different possible matches can be. If a very large value is used then anything can match anything else, which is unlikely to be useful.
The interface for the automatic remaps is as below. There are panes listing exact matches, non-matches (i.e. differences were too great), punctuation matches and possible typos. Users can choose to ignore any of these sets, and also select subsets within the sets if, for example, there are false matches.
 |
| The menus for Basedata, Matrices and Trees all have remap options, but there is also a centralised system under the File menu. |
 |
| Any object can be matched with any other object, as well as loading remaps from files. |
 |
| The Maximum acceptable distance controls how different names can be before they are no longer considered as possible matches. |
 |
| Users can control the level of remapping used within the match categories. |
Users can also choose to export the remap to a file to re-use later on (or inspect for possible problems) using the Export remap to file button. A copy of the remap can also be sent to the clipboard for direct use in spreadsheets, text editors and the like.
 |
| Remapping of BaseData objects is not permitted if they have existing outputs, as that would potentially wreak havoc with the matches between the BaseData and the outputs. |
 |
| The system will also warn if you try to map an object to itself (but it will let you try if you are determined to do so). |
Kudos to Luke Fitzpatrick who got it all working.
Shawn Laffan
26-Apr-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Constrain the extent of your exported results
This one is a short post.
Following a suggestion by Chris Barratt, Biodiverse version 2 will allow users to use a definition query when exporting spatial outputs. This means you can run your analysis for a large data set, but limit the set of exported groups to a subset.
If the analysis itself used a definition query then the export windows will set that by default. Just delete it to export all records.
Following a suggestion by Chris Barratt, Biodiverse version 2 will allow users to use a definition query when exporting spatial outputs. This means you can run your analysis for a large data set, but limit the set of exported groups to a subset.
If the analysis itself used a definition query then the export windows will set that by default. Just delete it to export all records.
 |
| Users can specify a definition query when exporting their data. In this case the analysis also used a definition query so it is specified by default. |
 |
| The exported data only contain those groups (cells) that passed the definition query. In this screenshot, no-data cells are in light grey to indicate the full extent of the exported data set. |
Shawn Laffan
26-Apr-2017
For more details about Biodiverse, see http://purl.org/biodiverse
For the full list of changes in the 1.99 series (leading to version 2) see https://purl.org/biodiverse/wiki/ReleaseNotes (for all issues addressed or being targeted to fix for version 2, see https://github.com/shawnlaffan/biodiverse/milestone/4 ).
To see what else Biodiverse has been used for, see https://purl.org/biodiverse/wiki/PublicationsList
You can also join the Biodiverse-users mailing list at http://groups.google.com/group/Biodiverse-users
Subscribe to:
Comments (Atom)